Gather files that fulfilled a given condition. Filter
[14]:
start_time = time.time()
# condition
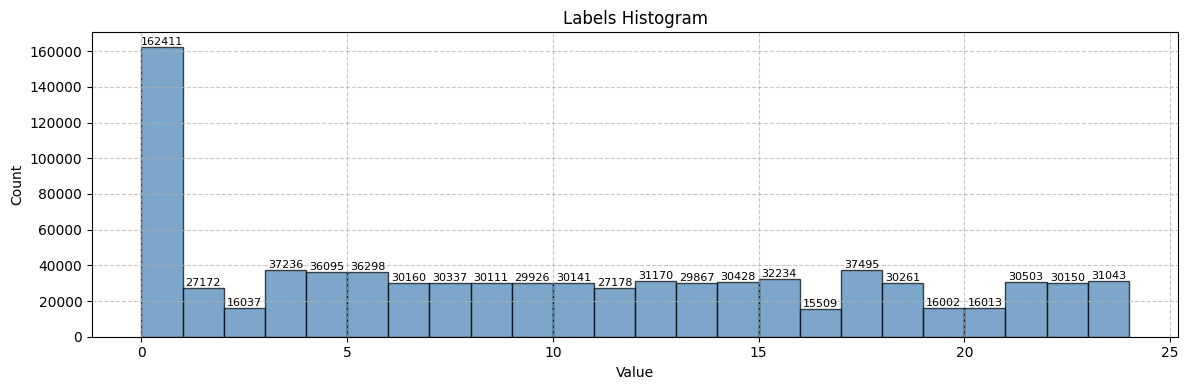
label_is_pipefittings = lambda ds: ds['face_labels'] == 15
filelist = explorer.get_file_list(group="faces", where=label_is_pipefittings)
print(f"Filtering completed in {(time.time() - start_time):.2f} seconds")
print(f"Found {len(filelist)} files with face_labels == 15 (chamfer)\n")
print(filelist)
Filtering completed in 0.21 seconds
Found 32234 files with face_labels == 15 (chamfer)
[ 10 12 15 ... 162392 162398 162401]
Query data for single file
[15]:
def demo_query_single_file(explorer, file_id):
"""Show how to access and query dataset details for a single file."""
print("=== Single File Dataset Access ===")
import time
# Get and print parquet info
df_info = explorer.get_parquet_info_by_code(file_id)
print("Files info:")
for column in df_info.columns:
print(f"Column: {column}")
for value in df_info[column]:
print(f" {value}")
print()
# Access various dataset groups
groups = ["faces", "file", "edges", "graph"]
datasets = {grp: explorer.file_dataset(file_id_code=file_id, group=grp) for grp in groups}
print(f"Datasets for file ID '{file_id}':")
for grp, ds in datasets.items():
for name, da in ds.data_vars.items():
print(f" [{grp}] VARIABLE: {name}, Shape: {da.shape}, Dims: {da.dims}, Size: {da.size}")
print()
# Query uv grids data for a specific face
start_time = time.time()
uv_grid_data = datasets["faces"]["face_uv_grids"].isel(face=2)
print("uv_grids data for face index 2:")
np_uvgrid = uv_grid_data.data.compute()
print(f"Query took {(time.time() - start_time):.2f} seconds\n")
[16]:
demo_query_single_file(explorer,file_id=4500)
=== Single File Dataset Access ===
Files info:
Column: name
07207dfd094fe0ebe9368ded4c271b23
Column: description
C:\Temp\Cadsynth_aag\step\20221124_154714_17096.step
Column: subset
test
Column: id
4500
Column: table_name
file_info
Datasets for file ID '4500':
[faces] VARIABLE: face_areas, Shape: (38,), Dims: ('face',), Size: 38
[faces] VARIABLE: face_indices, Shape: (38,), Dims: ('face',), Size: 38
[faces] VARIABLE: face_labels, Shape: (38,), Dims: ('face',), Size: 38
[faces] VARIABLE: face_loops, Shape: (38,), Dims: ('face',), Size: 38
[faces] VARIABLE: face_neighborscount, Shape: (38,), Dims: ('face',), Size: 38
[faces] VARIABLE: face_types, Shape: (38,), Dims: ('face',), Size: 38
[faces] VARIABLE: face_uv_grids, Shape: (38, 5, 5, 7), Dims: ('face', 'uv_x', 'uv_y', 'component'), Size: 6650
[faces] VARIABLE: file_id_code_faces, Shape: (38,), Dims: ('face',), Size: 38
[file] VARIABLE: duration_dglconvert, Shape: (1,), Dims: ('file',), Size: 1
[file] VARIABLE: file_id_code_file, Shape: (1,), Dims: ('file',), Size: 1
[file] VARIABLE: size_cadfile, Shape: (1,), Dims: ('file',), Size: 1
[file] VARIABLE: size_dglfile, Shape: (1,), Dims: ('file',), Size: 1
[edges] VARIABLE: edge_convexities, Shape: (96,), Dims: ('edge',), Size: 96
[edges] VARIABLE: edge_dihedral_angles, Shape: (96,), Dims: ('edge',), Size: 96
[edges] VARIABLE: edge_indices, Shape: (96,), Dims: ('edge',), Size: 96
[edges] VARIABLE: edge_lengths, Shape: (96,), Dims: ('edge',), Size: 96
[edges] VARIABLE: edge_types, Shape: (96,), Dims: ('edge',), Size: 96
[edges] VARIABLE: edge_u_grids, Shape: (96, 5, 6), Dims: ('edge', 'dim_x', 'component'), Size: 2880
[edges] VARIABLE: file_id_code_edges, Shape: (96,), Dims: ('edge',), Size: 96
[graph] VARIABLE: destination, Shape: (96,), Dims: ('edge',), Size: 96
[graph] VARIABLE: file_id_code_graph, Shape: (96,), Dims: ('edge',), Size: 96
[graph] VARIABLE: num_nodes, Shape: (96,), Dims: ('edge',), Size: 96
[graph] VARIABLE: source, Shape: (96,), Dims: ('edge',), Size: 96
uv_grids data for face index 2:
Query took 1.13 seconds
Create subsets (train, validation, test) based on the label distribution
[17]:
def demo_stratified_splits(explorer):
"""Show building a membership matrix and performing stratified splits."""
print("=== Membership Matrix and Data Splitting ===")
import time
from iterstrat.ml_stratifiers import MultilabelStratifiedShuffleSplit
import numpy as np
start_time = time.time()
df_label = explorer.get_descriptions("label", None, True)
label_key = df_label["table_name"].iloc[0]
if label_key == "file_label":
group = "file"
else:
group = "faces"
matrix, file_codes, _ = explorer.build_membership_matrix(group=group, key=label_key, bins_or_categories=None, as_counts=False)
# First split: 70% train, 30% temporary
msss = MultilabelStratifiedShuffleSplit(n_splits=1, test_size=0.30, random_state=42)
for train_idx, temp_idx in msss.split(np.arange(len(matrix))[:, None], matrix):
pass
# Second split on the temporary set into 50% validation, 50% test => 15% each overall
msss2 = MultilabelStratifiedShuffleSplit(n_splits=1, test_size=0.5, random_state=43)
for val_sub, test_sub in msss2.split(np.arange(len(temp_idx))[:, None], matrix[temp_idx]):
val_idx = temp_idx[val_sub]
test_idx = temp_idx[test_sub]
print("Train file IDs:", file_codes[train_idx].shape)
print("Validation file IDs:", file_codes[val_idx].shape)
print("Test file IDs:", file_codes[test_idx].shape)
print(f"Stratified Splitting completed in {(time.time() - start_time):.2f} seconds")
print()
[18]:
demo_stratified_splits(explorer)
=== Membership Matrix and Data Splitting ===
Train file IDs: (113479,)
Validation file IDs: (24453,)
Test file IDs: (24480,)
Stratified Splitting completed in 15.14 seconds