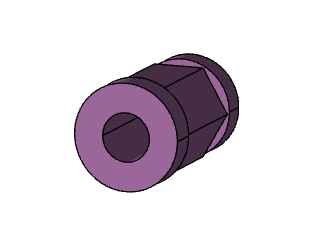
CAD Data Encoding
Overview
What is CAD Encoding?
The CAD Encoding module (
hoops_ai.cadencoder) is the bridge between CAD geometry and machine learning. It transforms symbolic CAD representations (surfaces, curves, topological relationships) into numeric feature vectors that neural networks can process.Tip
New to CAD or ML? This guide assumes familiarity with:
CAD concepts: B-rep, Topology, faces, edges → See CAD Fundamentals
ML concepts: Feature vectors, graph neural networks, node features → See Machine Learning Fundamentals
Key terms: Check the Glossary for quick definitions
The Core Challenge:
CAD files store geometry in a semantic, mathematical form (symbolic representations with precise meaning):
A planar face is stored as: “Plane equation: \(0.707x + 0.707y + 0z = 10\)”
A cylindrical face is: “Cylinder with axis \([0, 0, 1]\), radius \(5mm\)”
Face adjacency is: “Face #5 and Face #12 share edge #23”
Machine learning models need fixed-size numeric vectors (feature vectors - arrays of numbers representing object properties):
Face features:
[area, perimeter, centroid_x, centroid_y, centroid_z, ...]Edge features:
[length, angle, curvature, ...]Graph structure:
edges = [[src_nodes], [dst_nodes]]
What This Module Does:
hoops_ai.cadencoderprovides theBrepEncoderclass that:
Queries CAD data using
HOOPSBrepinterface (fromcadaccessmodule)Computes numeric features (areas, lengths, angles, surface types)
Structures data for ML (face adjacency graphs, feature arrays, UV grids)
Persists to storage using
DataStorageinterface (fromstoragemodule)Why “Push” Methods?
The
BrepEncoderclass computes and persists geometric and topological features from BREP data. It follows a push-based architecture where each method:
Checks if data already exists in storage
Ensures the appropriate schema definition exists for the data
Computes the feature if needed
Saves to storage with schema management
Returns None (if storage is used) or the computed data (if no storage)
The encoder automatically manages schemas for data organization, creating groups and arrays as needed during the encoding process.
See also
CAD Data Access - How to query CAD geometry using HOOPSBrep
Data Storage - How DataStorage works and when to use different backends
CAD Fundamentals - Understanding B-rep topology and geometry
Architecture - How Encoding Works
The Data Flow:
CAD File (part.step) ↓ HOOPSLoader (loads file) ↓ HOOPSModel (in-memory representation) ↓ HOOPSBrep (query interface) ↓ BrepEncoder ←--------→ DataStorage (feature extraction) (persistence) ↓ Encoded Dataset (.data file or memory dict)Component Interaction:
HOOPSBrep: Provides query methods (
get_face_attributes(),get_edge_attributes(),build_face_adjacency_graph(), etc.)BrepEncoder: Orchestrates feature extraction by calling
HOOPSBrepmethodsDataStorage: Receives extracted features and saves them (Zarr arrays, JSON, etc.)
Why This Architecture?:
Separation of Concerns:
HOOPSBrepfocuses on queries, BrepEncoder on feature engineering,DataStorageon persistenceTestability: Mock
HOOPSBrepandDataStoragefor unit testing encodersFlexibility: Swap storage backends without changing encoding logic
Tip
Getting Started? If you’re new to this workflow:
Start with the simple example in the next section
See Tutorials for hands-on encoding walkthroughs
Understand what gets encoded by reading CAD Fundamentals first
The BrepEncoder Class
What is BrepEncoder?
BrepEncoder is the main feature extraction engine in HOOPS AI. It systematically processes a B-rep model and generates all the numeric features needed for ML training.
Initialization:
from hoops_ai.cadencoder import BrepEncoder from hoops_ai.storage import DataStorage # With storage storage = DataStorage(...) encoder = BrepEncoder(brep_access=brep, storage_handler=storage) # Without storage (returns raw data) encoder = BrepEncoder(brep_access=brep)Parameters:
brep_access (HOOPSBrep): BREP interface from a loaded CAD model
storage_handler (DataStorage, optional): Storage backend for persistence
Constructor Signature:
def __init__(self, brep_access: HOOPSBrep, storage_handler: DataStorage = None): """ Args: brep_access: The B-Rep geometry data source interface (from cadaccess module) storage_handler: Optional, object to load/save data from disk or memory """What the Encoder Needs:
A
hoops_ai.cadaccess.HOOPSBrepobject (from loaded CAD model) - this is the “source” of geometric queriesOptional
hoops_ai.storage.DataStorageobject - this is the storage object where features are saved (if None, methods return data directly)
Note
Prerequisite: Understand what face adjacency graphs are and why they matter. See:
CAD Fundamentals - B-rep Topology section explains faces, edges, adjacency
CAD Data Access - Topological Queries section shows how to build adjacency graphs
Topology Encoding Methods
What is Topology Encoding?
Topology encoding extracts the connectivity structure of the B-rep - which entities connect to which. This is distinct from geometry encoding (sizes, shapes, positions). See CAD Fundamentals for the difference between Topology and geometry.
Why Topology Matters for ML:
Graph Neural Networks: Topology defines the graph edges (Message Passing paths) - see Machine Learning Fundamentals for how GNNs use graph structure
Feature Recognition: Machining features are subgraphs with specific topology (e.g., “pocket = 6 connected planar faces forming a box”)
Manufacturing Constraints: Adjacent faces must have compatible machining directions
Segmentation: Group faces that are topologically connected
BrepEncoder.push_face_adjacency_graph()
The method BrepEncoder.push_face_adjacency_graph builds a face adjacency graph from the B-rep model. This graph represents the topology of the model where nodes are faces and edges connect adjacent faces.
What It Does:
Build a graph representation of face connectivity where faces are nodes and edges represent shared boundaries.
Mathematical Formulation:
Define an undirected graph \(G=(V,E)\) where:
\[ \begin{align}\begin{aligned}V = \mathcal{F} = \{f_0, f_1, \ldots, f_{N_f-1}\}\\E = \{(f_i, f_j) : f_i \text{ and } f_j \text{ share an edge}\}\end{aligned}\end{align} \]The graph is represented by:
Node count: \(|V| = N_f\)
Edge list: \(\{(s_k, d_k)\}_{k=0}^{|E|-1}\) where \(s_k, d_k \in V\)
Method Signature:
def push_face_adjacency_graph(self) -> Union[Tuple[str, int, int], nx.Graph]: """ Returns: If storage_handler is not None: Tuple[str, int, int] - (storage_key, num_faces, num_edges) If storage_handler is None: nx.Graph - the face adjacency graph directly """Usage:
# we assume 'cad_model' is your loaded CADModel instance from hoops_ai.cadencoder import BrepEncoder brep_encoder = BrepEncoder(cad_model.get_brep()) adj_graph = brep_encoder.push_face_adjacency_graph() print(adj_graph) import networkx as nx import matplotlib.pyplot as plt pos = nx.spring_layout(adj_graph) # compute layout once nx.draw_networkx(adj_graph, pos, arrows=False) # draw nodes, edges, labels plt.axis('off') # turn off axes for clarity plt.show()Example Output:
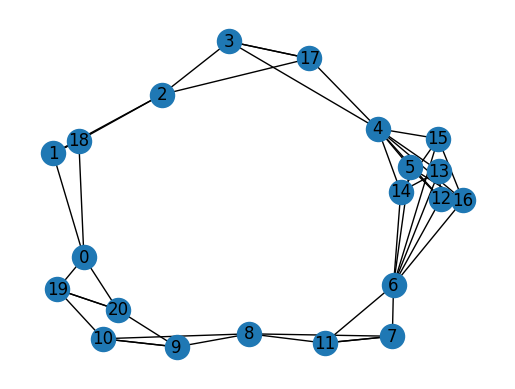
This example shows a
DiGraph with 21 nodes and 46 edges. The images below show the 3D CAD model and its corresponding face adjacency graph:The graph visualization shows nodes (faces) numbered 0-20 and edges connecting adjacent faces. The spring layout algorithm positions the nodes for clarity. Each node in the graph corresponds to a face in the 3D model, and edges represent shared boundaries between faces.
Storage Format Details:
The encoder stores graph data in two formats simultaneously for compatibility:
Flat arrays:
num_nodes: scalar count of nodes in the graph
edges_source: source node indices for each edge
edges_destination: destination node indices for each edge
graph: nested structure containing edges dict and num_nodes (for backward compatibility)
Nested dictionary: Dtypes: int32
Returns:
With storage: Returns
None(data is stored with keys: “num_nodes”, “edges_source”, “edges_destination”, and “graph”)Without storage: Returns
nx.Graph- NetworkX graph object with edge attributes
Note
Understanding the format: If you’re unfamiliar with graph representations for ML, see Machine Learning Fundamentals - the “Graph Representation of CAD Models” section explains nodes, edges, and node features.
BrepEncoder.push_extended_adjacency()
The method BrepEncoder.push_extended_adjacency computes the extended adjacency matrix representing shortest path distances between all pairs of faces and ensures ‘extended_adjacency’ is in storage or returns the extended adjacency data directly.
What It Does:
Computes shortest path distances between all pairs of faces using Floyd-Warshall algorithm on the face adjacency graph. This provides global topological context.
Method Signature:
def push_extended_adjacency(self) -> Union[str, np.ndarray]:
"""
Returns:
If storage_handler is not None: str - storage key ("extended_adjacency")
If storage_handler is None: np.ndarray - shape (num_faces, num_faces)
"""
Usage:
# Compute all-pairs shortest paths key = encoder.push_extended_adjacency() # Later: check topological distance distances = storage.load_data("extended_adjacency") # distances[i, j] = shortest path length from face i to face j # distances[i, i] = 0 (same face) # distances[i, j] = 1 (directly adjacent) # distances[i, j] = 2 (connected through one intermediate face)
Mathematical Formulation:
Compute the graph distance matrix \(\mathbf{D}_G \in \mathbb{R}^{N_f \times N_f}\):
\[\begin{split}D_G[i,j] = \begin{cases} 0 & \text{if } i = j \\ \min\{|p| : p \text{ is path from } f_i \text{ to } f_j\} & \text{if path exists} \\ \infty & \text{otherwise} \end{cases}\end{split}\]
where \(|p|\) is the number of edges in path \(p\).
This is computed using the Floyd-Warshall or BFS algorithm via NetworkX’s all_pairs_shortest_path_length.
Storage:
Array: extended_adjacency
Shape: [node_i, node_j]
Dtype: float32
Returns:
With storage: Returns
None(data is stored with key"extended_adjacency")Without storage:
np.ndarrayof shape(N_f, N_f)
BrepEncoder.push_face_neighbors_count()
The method BrepEncoder.push_face_neighbors_count counts the number of adjacent faces for each face and ensures ‘face_neighborscount’ is in storage or returns the neighbor counts directly.
What It Does:
Count the number of adjacent faces for each face (node degree in the graph).
Usage:
key = encoder.push_face_neighbors_count() neighbor_counts = storage.load_data("face_neighborscount") # neighbor_counts[i] = number of faces adjacent to face i
Mathematical Formulation:
For each face \(f_i\), compute the degree:
\[\deg(f_i) = |\{f_j \in \mathcal{F} : (f_i, f_j) \in E\}|\]
- Storage:
Array:
face_neighborscountShape:
[face]Dtype:
int32
- Returns:
With storage: Returns
None(data is stored with key"face_neighborscount")Without storage:
np.ndarrayof shape(N_f,)
BrepEncoder.push_face_pair_edges_path(max_allow_edge_length=16)
The method BrepEncoder.push_face_pair_edges_path computes the sequence of edges along the shortest path between all pairs of faces and ensures ‘face_pair_edges_path’ is in storage or returns the edge paths directly.
What It Does:
Store the sequence of shared edges along the shortest path between every pair of faces.
Usage:
key = encoder.push_face_pair_edges_path(max_allow_edge_length=16) edge_paths = storage.load_data("face_pair_edges_path") # Shape: (num_faces, num_faces, 16) # edge_paths[i, j, :] = edge indices from face i to face j (-1 for padding)
Mathematical Formulation:
For each face pair \((f_i, f_j)\), find the shortest path:
\[p_{ij} = [f_i = v_0, v_1, \ldots, v_k = f_j]\]Then extract the edge sequence:
\[\mathbf{e}_{ij} = [e(v_0, v_1), e(v_1, v_2), \ldots, e(v_{k-1}, v_k)]\]where \(e(u,v)\) is the edge index connecting faces \(u\) and \(v\) .
If \(|\mathbf{e}_{ij}| > M\) (max_allow_edge_length), truncate to first \(M\) edges.
Pad with \(-1\) if path is shorter.
Storage:
Array:
face_pair_edges_pathShape:
[face_i, face_j, path_idx]Dtype:
int32
Parameters:
max_allow_edge_length (int): Maximum path length to store (default: 16)
Returns:
With storage: Returns
None(data is stored with key"face_pair_edges_path")Without storage:
np.ndarrayof shape(N_f, N_f, M)
Geometry Encoding Methods
What is Geometry Encoding?
Geometry encoding extracts numeric measurements of CAD entities - sizes, shapes, positions, and curvatures. While Topology tells us which faces are connected, geometry tells us their actual physical properties. See CAD Fundamentals for the topology vs. geometry distinction.
BrepEncoder.push_face_attributes()
The method BrepEncoder.push_face_attributes Extracts and stores various face attributes, including face types, areas, and loop counts.
What It Does:
Compute geometric and topological properties of each face.
Method Signature:
def push_face_attributes(self) -> Union[Tuple[List[str], Dict], Tuple[List[np.ndarray], Dict]]:
"""
Returns:
If storage_handler is not None:
Tuple[List[str], Dict] - (list_of_stored_keys, face_type_descriptions)
Example: (['face_types', 'face_areas', 'face_loops'], {9: 'Plane', 10: 'Cylinder'})
If storage_handler is None:
Tuple[List[np.ndarray], Dict] - (list_of_arrays, face_type_descriptions)
"""
Usage:
With Storage Handler:
# Extract face attributes (with storage handler) keys, face_type_desc = encoder.push_face_attributes() print(f"Stored face data at keys: {keys}") # Output: ['face_types', 'face_areas', 'face_loops'] print(f"Face type descriptions:") for type_id, description in face_type_desc.items(): print(f" {type_id}: {description}") # Output: # 9: Plane # 10: Cylinder # 11: Cone # ... # Later: retrieve from storage face_types = storage.load_data("face_types") # int32 array[num_faces] face_areas = storage.load_data("face_areas") # float32 array[num_faces] face_loops = storage.load_data("face_loops") # int32 array[num_faces]
- Without Storage Handler:
Example Output:
face_types [0 1 0 1 0 0 0 1 0 1 1 1 0 0 0 0 0 1 1 2 2] face_areas [ 43.911655 141.3149 75.277115 51.831074 24.732485 57.030937 19.963306 12.871587 28.228918 39.265965 39.265965 12.871587 57.030937 57.030933 57.030937 57.030937 57.030933 51.831074 141.3149 10.575602 10.575602] face_loops [2 1 2 1 2 1 2 1 2 1 1 1 1 1 1 1 1 1 1 1 1] face_types_descr {0: 'Plane', 1: 'Cylinder', 2: 'Cone'}
Mathematical Formulation:
For each face \(f_i \in \mathcal{F}\):
Surface Type \(\tau(f_i)\): Categorical classification (plane, cylinder, sphere, etc.)
\[\tau: \mathcal{F} \rightarrow \mathbb{Z}^+\]
Face Area \(A(f_i)\): Surface integral over the face
\[A(f_i) = \iint_{S_i} dS\]
Loop Count \(L(f_i)\): Number of boundary loops (including holes)
\[L(f_i) = |\{\text{loops in } f_i\}|\]
- Storage:
Arrays:
face_types,face_areas,face_loopsShapes: All
[face]Dtypes:
int32,float32,int32
Returns:
With storage: Returns
None(data is stored with keys:"face_types","face_areas","face_loops", and metadata"descriptions/face_types")Without storage:
Tuple[List[np.ndarray], Dict]- (list of numpy arrays [face_types, face_areas, face_loops], face_types_descr dictionary mapping face type IDs to descriptions)
BrepEncoder.push_edge_attributes()
The method
BrepEncoder.push_edge_attributesExtracts and stores various edge attributes, including edge types, lengths, dihedral angles, and convexities.What It Does:
Compute geometric and topological properties of each edge.
Method Signature:
def push_edge_attributes(self) -> Union[Tuple[List[str], Dict], Tuple[List[np.ndarray], Dict]]: """ Returns: If storage_handler is not None: Tuple[List[str], Dict] - (list_of_stored_keys, edge_type_descriptions) If storage_handler is None: Tuple[List[np.ndarray], Dict] - (list_of_arrays, edge_type_descriptions) """Mathematical Formulation:
For each edge \(e_i \in \mathcal{E}\):
Curve Type \(\kappa(e_i)\): Categorical classification (line, circle, spline, etc.)
\[\kappa: \mathcal{E} \rightarrow \mathbb{Z}^+\]
Edge Length \(\ell(e_i)\): Arc length of the curve
\[\ell(e_i) = \int_{0}^{1} \left\| \frac{d\mathbf{C}(t)}{dt} \right\| dt\]where \(\mathbf{C}(t)\) is the curve parameterization.
Dihedral Angle \(\theta(e_i)\): Angle between adjacent face normals
\[\theta(e_i) = \arccos(\mathbf{n}_1 \cdot \mathbf{n}_2)\]where \(\mathbf{n}_1, \mathbf{n}_2\) are unit normals of adjacent faces.
Convexity \(\chi(e_i) \in \{-1, 0, 1\}\):
\[\begin{split}\chi(e_i) = \begin{cases} 1 & \text{if convex} \\ 0 & \text{if smooth} \\ -1 & \text{if concave} \end{cases}\end{split}\]
Storage:
Arrays:
edge_types,edge_lengths,edge_dihedral_angles,edge_convexitiesShapes: All
[edge]Dtypes:
int32,float32,float32,int32
Returns:
With storage: Returns
None(data is stored with keys:"edge_types","edge_lengths","edge_dihedral_angles","edge_convexities", and metadata"descriptions/edge_types")Without storage:
Tuple[List[np.ndarray], Dict]- (list of numpy arrays [edge_types, edge_lengths, edge_dihedrals, edge_convexities], edge_type_descrip dictionary mapping edge type IDs to descriptions)
Usage:
With Storage Handler:
# Extract edge attributes (with storage handler) keys, edge_type_desc = encoder.push_edge_attributes() print(f"Stored edge data at keys: {keys}") # Output: ['edge_types', 'edge_lengths', 'edge_dihedral_angles', 'edge_convexities'] # Later: retrieve from storage edge_types = storage.load_data("edge_types") # int32 array[num_edges] edge_lengths = storage.load_data("edge_lengths") # float32 array[num_edges] edge_dihedrals = storage.load_data("edge_dihedral_angles") # float32 array[num_edges] edge_convexities = storage.load_data("edge_convexities") # int32 array[num_edges]
- Without Storage Handler:
Example Output:
edge_types_np [1 1 1 1 0 1 0 1 1 1 0 0 1 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 1 0 1 0 1 1 1 0 0 1 1 0 0 0 0 0 0] edge_lengths_np [14.137167 14.137167 7.8539815 7.8539815 18. 7.8539815 18. 17.278759 17.278759 7.8539815 3. 3. 17.278759 17.278759 5.196152 5.196152 5.196152 5.196152 5.196152 5.196152 10.97561 5.196152 10.97561 5.196152 5.196152 5.196152 5.196152 5.196152 12.566371 12.566371 1.0243902 12.566371 1.0243902 15.707963 15.707963 12.566371 2.5 2.5 15.707963 15.707963 10.97561 10.97561 10.97561 10.97561 0.70710677 0.70710677] edge_dihedrals_np [ 7.8539819e-01 7.8539819e-01 -1.5707964e+00 -1.5707964e+00 2.4492937e-16 1.5707964e+00 0.0000000e+00 -1.5707964e+00 -1.5707964e+00 1.5707964e+00 0.0000000e+00 2.4492937e-16 1.5707964e+00 1.5707964e+00 -1.5707964e+00 -1.5707964e+00 -1.5707964e+00 -1.5707964e+00 -1.5707964e+00 -1.5707964e+00 1.0471976e+00 1.5707964e+00 1.0471976e+00 1.5707964e+00 1.5707964e+00 1.5707964e+00 1.5707964e+00 1.5707964e+00 -1.5707964e+00 -1.5707964e+00 2.4492937e-16 1.5707964e+00 0.0000000e+00 -1.5707964e+00 -1.5707964e+00 1.5707964e+00 0.0000000e+00 2.4492937e-16 7.8539819e-01 7.8539819e-01 1.0471976e+00 1.0471976e+00 1.0471976e+00 1.0471976e+00 0.0000000e+00 0.0000000e+00] edge_convexities_np [1 1 2 2 3 1 3 2 2 1 3 3 1 1 2 2 2 2 2 2 1 1 1 1 1 1 1 1 2 2 3 1 3 2 2 1 3 3 1 1 1 1 1 1 3 3] edge_type_descrip {1: 'Circle', 0: 'Line'}
What Gets Stored:
Storage Key
Type
Shape
Description
edge_typesint32
(num_edges,)Curve type IDs (1=Line, 2=Circle, 3=Ellipse, 4=NURBS, etc.)
edge_lengthsfloat32
(num_edges,)Curve length in model units
edge_dihedral_anglesfloat32
(num_edges,)Angle between adjacent faces (radians)
edge_convexitiesint32
(num_edges,)Convexity: 1=convex, -1=concave, 0=smooth/tangent
descriptions/edge_typesmetadata
dict
Human-readable curve type names:
{1: 'Line', 2: 'Circle', ...}
BrepEncoder.push_curvegrid(ugrid=5)
The method BrepEncoder.push_curvegrid Samples points along edges at regular intervals.
What It Does:
Sample points and tangents along edge curves.
Method Signature:
def push_curvegrid(self, ugrid: int = 5) -> Union[str, np.ndarray]: """ Args: ugrid: Number of samples along each edge (default: 5) Returns: If storage_handler is not None: str - storage key ("edge_u_grids") If storage_handler is None: np.ndarray - shape (num_edges, ugrid-2, 6) """
Usage:
edge_grids = brep_encoder.push_curvegrid(ugrid=3) print("edge_grids\n", edge_grids[0])
Example Output:
edge_grids [[ 2.2000000e+01 4.5000000e+00 0.0000000e+00 -0.0000000e+00 0.0000000e+00 -1.0000000e+00] [ 2.2000000e+01 2.2500000e+00 -3.8971143e+00 0.0000000e+00 -8.6602539e-01 -5.0000000e-01] [ 2.2000000e+01 -4.5000000e+00 -1.5436120e-14 0.0000000e+00 -3.4302490e-15 1.0000000e+00]]
Mathematical Formulation:
For each edge \(e_i\), sample along the curve parameter:
where:
\(\mathbf{C}: [0,1] \rightarrow \mathbb{R}^3\) is the curve
\(\mathbf{T}(t) = \frac{d\mathbf{C}(t)}{dt}\) is the tangent vector
\(t_j = \frac{j}{U-1}\) for \(j = 0, \ldots, U-1\)
Storage:
Array:
edge_u_gridsShape:
[edge, u, component]where component includes (x,y,z) + (tx,ty,tz)Dtype:
float32
Parameters:
ugrid(int): Number of samples along edge (default: 5)
Returns:
With storage:
str- key name"edge_u_grids"Without storage:
np.ndarrayof shape(N_e, ugrid, 6)
BrepEncoder.push_face_indices()
What it does: Extract and store unique identifiers for all faces in the model.
Mathematical Formulation:
where \(\mathcal{F}\) is the set of face indices and \(N_f\) is the total number of faces.
Storage:
Group:
facesArray:
face_indicesShape:
[face]Dtype:
int32
Returns:
With storage:
str- key name"face_indices"Without storage:
np.ndarrayof shape(N_f,)
BrepEncoder.push_edge_indices()
What it does: Extract and store unique identifiers for all edges in the model.
Mathematical Formulation:
where \(\mathcal{E}\) is the set of edge indices and \(N_e\) is the total number of edges.
Storage:
Group:
edgesArray:
edge_indicesShape:
[edge]Dtype:
int32
Returns:
With storage:
str- key name"edge_indices"Without storage:
np.ndarrayof shape(N_e,)
BrepEncoder.push_face_discretization(pointsamples=25)
What it does: Sample points and normals on face surfaces using uniform point sampling (rather than structured UV grids).
Mathematical Formulation:
For each face \(f_i\), sample \(P\) points uniformly across the surface:
where:
\(\mathbf{S}: \Omega \rightarrow \mathbb{R}^3\) is the surface parameterization
\(\mathbf{N}: \Omega \rightarrow \mathbb{S}^2\) is the normal field
\(V: \Omega \rightarrow \{0,1\}\) is visibility status (inside/outside)
\(\mathbf{u}_j\) are uniformly sampled parameter points across the face
\(P\) is the number of sample points (default: 25)
The sampling uses three methods concatenated along the component axis:
Point samples: \((x, y, z)\) coordinates
Normal samples: \((n_x, n_y, n_z)\) unit normals
Inside/outside flags: visibility indicators
Storage:
Array:
face_discretizationShape:
[face, sample, component]where component includes (x,y,z) + (nx,ny,nz) + (visibility)Dtype:
float32
Parameters:
pointsamples(int): Number of points to sample per face (default: 25)
Returns:
With storage:
str- key name"face_discretization"Without storage:
np.ndarrayof shape(N_f, pointsamples, 7)
Histogram-Based Features
BrepEncoder.push_average_face_pair_distance_histograms(grid=5, num_bins=64)
The method BrepEncoder.push_average_face_pair_distance_histograms computes histograms of point-to-point distances between all pairs of faces and ensures ‘d2_distance’ is in storage or returns the distance histograms directly.
What It Does:
Compute normalized histograms of pairwise point-to-point distances between all face pairs (D2 shape descriptor).
Usage:
key = encoder.push_average_face_pair_distance_histograms(grid=5, num_bins=64) distance_histograms = storage.load_data("d2_distance") # Shape: (num_faces, num_faces, 64) # Histogram of distances between sample points from face i and face j
Implementation Notes:
Uses optimized sampling: maximum 25 points per face (or fewer if face has less than 25 points)
Employs 2-thread parallel processing for improved performance
Processes faces in two chunks to balance memory and computation
Mathematical Formulation:
Sample Points: For each face \(f_i\), sample \(P\) points uniformly:
Compute Distances: For faces \(f_i\) and \(f_j\), compute all pairwise distances:
Normalize by Diagonal: Let \(D\) be the bounding box diagonal:
Normalized distances:
Build Histogram: Bin the normalized distances into \(B\) bins over \([0,1]\):
Result: \(\mathbf{H} \in \mathbb{R}^{N_f \times N_f \times B}\) where \(H_{ij}\) is the distance histogram between faces \(i\) and \(j\).
Storage:
Group:
histogramsArray:
d2_distanceShape:
[face_i, face_j, bin]Dtype:
float32
Parameters:
grid(int): Grid density for sampling (default: 5)
num_bins(int): Number of histogram bins (default: 64)
Returns:
With storage: Returns
None(data is stored with key"d2_distance")Without storage:
np.ndarrayof shape(N_f, N_f, num_bins)
BrepEncoder.push_average_face_pair_angle_histograms(grid=5, num_bins=64)
The method BrepEncoder.push_average_face_pair_angle_histograms computes histograms of angles between normals for all pairs of faces and ensures ‘a3_distance’ is in storage or returns the angle histograms directly.
What It Does:
Compute normalized histograms of pairwise normal-to-normal angles between all face pairs (A3 shape descriptor).
Implementation Notes:
Uses optimized sampling: maximum 25 normals per face (or fewer if face has less than 25 normals)
Employs 2-thread parallel processing for improved performance
Processes faces in two chunks to balance memory and computation
Usage:
key = encoder.push_average_face_pair_angle_histograms(grid=5, num_bins=64) angle_histograms = storage.load_data("a3_distance") # Shape: (num_faces, num_faces, 64) # Histogram of angles between normal vectors from face i and face j
Mathematical Formulation:
Sample Normals: For each face \(f_i\), sample \(P\) normal vectors:
\[\mathcal{N}_i = \{\mathbf{n}_1^i, \mathbf{n}_2^i, \ldots, \mathbf{n}_P^i\} \subset \mathbb{S}^2\]Compute Angles: For faces \(f_i\) and \(f_j\), compute all pairwise angles:
\[\theta_{ij}^{mn} = \arccos(\mathbf{n}_m^i \cdot \mathbf{n}_n^j), \quad m,n = 1,\ldots,P\]
Clamping: \(\mathbf{n}_m^i \cdot \mathbf{n}_n^j \in [-1, 1]\) to avoid numerical issues.
Normalize to [0,1]:
\[\tilde{\theta}_{ij}^{mn} = \frac{\theta_{ij}^{mn}}{\pi}\]Build Histogram: Bin the normalized angles into \(B\) bins:
\[H_{ij}^{\theta}[b] = \frac{1}{P^2} \sum_{m=1}^P \sum_{n=1}^P \mathbb{1}\left[\frac{b}{B} \leq \tilde{\theta}_{ij}^{mn} < \frac{b+1}{B}\right]\]
Result: \(\mathbf{H}^{\theta} \in \mathbb{R}^{N_f \times N_f \times B}\) where \(H_{ij}^{\theta}\) is the angle histogram between faces \(i\) and \(j\).
Storage:
Group:
histogramsArray:
a3_distanceShape:
[face_i, face_j, bin]Dtype:
float32
Parameters:
grid(int): Grid density for sampling normals (default: 5)
num_bins(int): Number of histogram bins (default: 64)
Returns:
With storage: Returns
None(data is stored with key"a3_distance")Without storage:
np.ndarrayof shape(N_f, N_f, num_bins)
Complete Encoding Example
Here’s a comprehensive encoding workflow following the actual usage pattern from the tutorials:
from hoops_ai.cadaccess import HOOPSLoader
from hoops_ai.cadencoder import BrepEncoder
from hoops_ai.storage import OptStorage
# 1. Load CAD file
loader = HOOPSLoader()
model = loader.create_from_file("part.step")
# 2. Extract BREP
brep = model.get_brep()
# 3. Initialize storage and encoder
storage = OptStorage(output_path="./encoded_data")
encoder = BrepEncoder(brep_access=brep, storage_handler=storage)
# 4. Extract geometric features
encoder.push_face_indices()
encoder.push_edge_indices()
encoder.push_face_attributes()
encoder.push_edge_attributes()
# 5. Extract parameterized grids
encoder.push_face_discretization(pointsamples=100)
encoder.push_curvegrid(ugrid=20)
# 6. Extract topology
encoder.push_face_adjacency_graph()
encoder.push_extended_adjacency()
encoder.push_face_neighbors_count()
# 7. Extract shape descriptors
encoder.push_average_face_pair_distance_histograms(grid=7, num_bins=64)
encoder.push_average_face_pair_angle_histograms(grid=7, num_bins=64)
print("Encoding complete!")
Performance Considerations
Memory Management
The encoder uses a push-and-discard pattern: data is computed, saved, and not kept in memory
Large arrays (histograms) use chunked processing with ThreadPoolExecutor
UV grids and curve grids are stacked only temporarily
Parallelization
Face pair histograms use 2-thread parallel processing
Sampling operations are vectorized with NumPy
Graph algorithms leverage NetworkX’s optimized implementations
Storage Efficiency
Float32 is used throughout for memory/disk efficiency
Zarr format provides compression and chunked access
Schema management ensures consistent data organization
Next Steps
Read Data Storage to understand DataStorage backends (OptStorage, MemoryStorage)
Study Datasets - ML-Ready Inputs for batch processing and dataset management
Review Data Flow Customisation for automating encoding across many CAD files
Try hands-on tutorials in Tutorials for practical examples